Research
We develop experimental methods for high-throughput microscopy and biochemical profiling of primary and human induced pluripotent stem cell (iPSC)-derived cardiomyocytes. Our computational approaches include large-scale regulatory network modeling and bioinformatic analysis of -omic data. Specific research focus areas include cardiomyocyte hypertrophy and death, extra-cellular matrix remodeling by fibroblasts and macrophages, and cardiomyocyte proliferation.
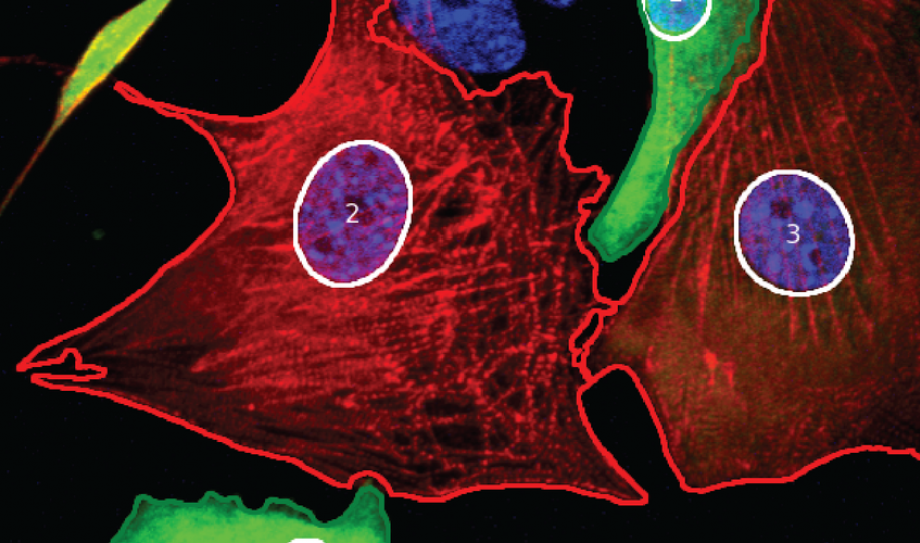
Automated image analysis of cardiac cells and tissue.
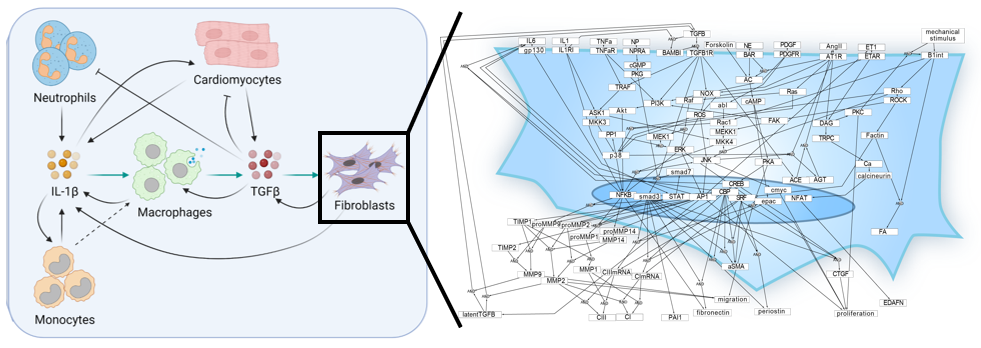
Multi-scale modeling of inflammation-fibrosis coupling
Selected Publications
Nelson AR, Christiansen SL, Naegle KM, Saucerman JJ.Proc Natl Acad Sci U S A. 2024
Chowkwale M, Lindsey ML, Saucerman JJ.J Physiol. 2023 Jul;601(13):2635-2654.
Virtual drug screen reveals context-dependent inhibition of cardiomyocyte hypertrophy.
Eggertsen TG, Saucerman JJ.Br J Pharmacol. 2023 Nov;180(21):2721-2735.
Brahma safeguards canalization of cardiac mesoderm differentiation.
Hota SK, Rao KS, Blair AP, Khalilimeybodi A, Hu KM, Thomas R, So K, Kameswaran V, Xu J, Polacco BJ, Desai RV, Chatterjee N, Hsu A, Muncie JM, Blotnick AM, Winchester SAB, Weinberger LS, Hüttenhain R, Kathiriya IS, Krogan NJ, Saucerman JJ, Bruneau BG.Nature. 2022 Feb;602(7895):129-134.
Publications
CSBG publications on PUBMED
Downloads
Our open-source software and computational models on GITHUB
You can find our data on FIGSHARE
Netflux: User-friendly software for modeling biological networks https://github.com/saucermanlab/Netflux
